IGV with all data produced by Scrimer¶
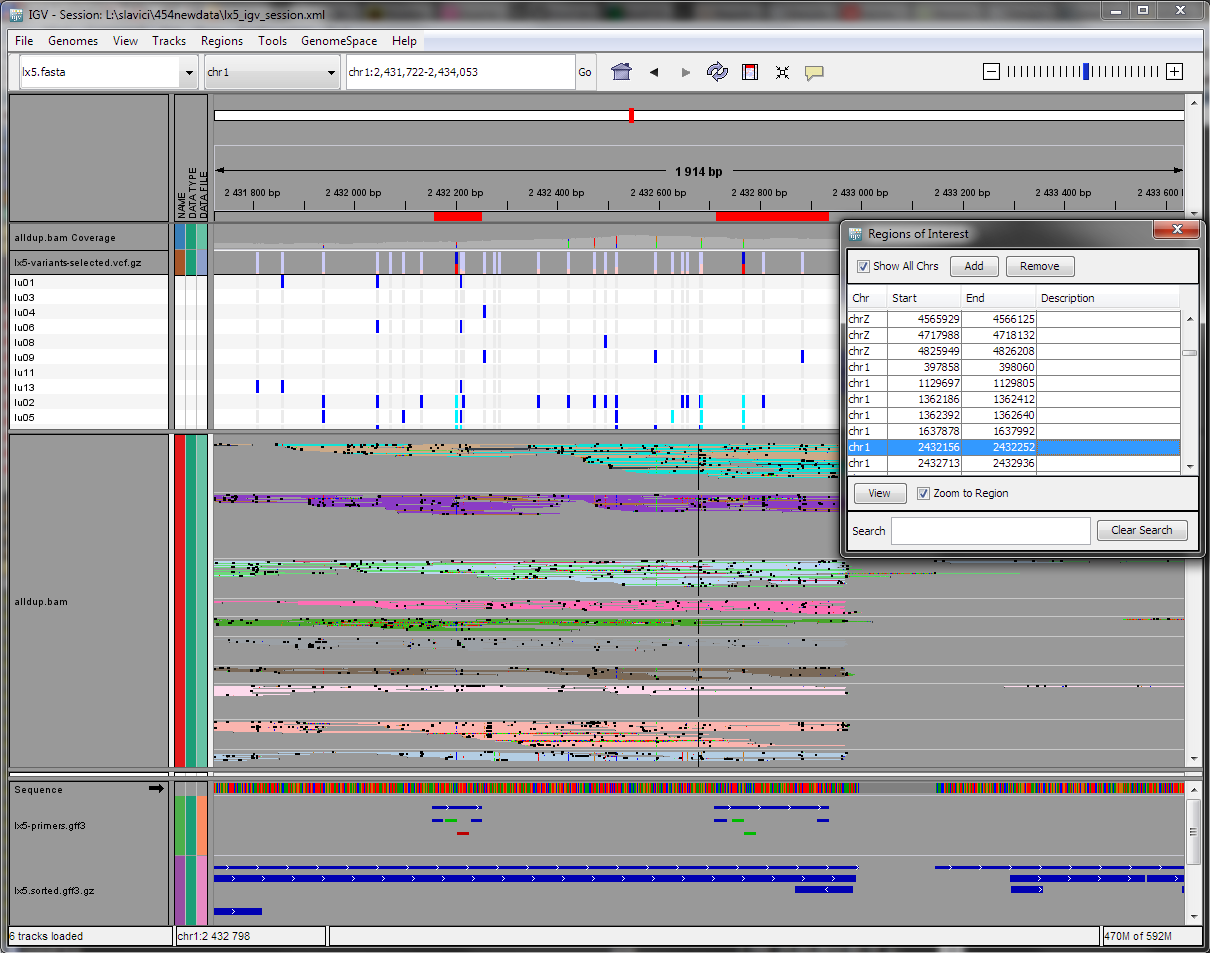
Tracks, from top to bottom:
- genome navigator, ‘genome’ produced in Map contigs to the reference genome
alldup.bam coverage- total coverage for the region, produced in Map reads to the scaffoldlx5-variants-selected.vcf.gz- summary of the variants, filtered variants are shown in lighter color, produced in Detect and choose variants- sample list - provides detailed information on variants in each sample
alldup.bam- details on coverage and SNP (colored) / INDEL (black), in the context menu chooseGroup alignments by > sampleandColor alignments by > read group, produced in Map reads to the scaffold- sequence
lx5-primers.gff3- resulting primers, hover with mouse for details on calculated properties, produced in Design primerslx5.sorted.gff3.gz- annotations for the scaffold - predicted and transferred exons, produced in Map contigs to the reference genome- floating window with a list of designed primers, produced in Design primers
How to get to this view¶
- run IGV (version 2.2 is used here)
Genomes > Load genome from file, choose your scaffold- load all the tracks by
File > Load from File - rearrange tracks according to your preference by dragging the label with the mouse
- choose
Regions > Import Regions, pick the.bedfile created in Design primers, chooseRegions > Region Navigator